I’m a PhD candidate in Computer Science at CUNY Graduate Center, where I build AI systems that decode biology and reimagine drug discovery—think virtual humans simulating how drugs work before trials begin. I’m advised by Dr. Lei Xie (The City University of New York & Weill Cornell), and my work spans large language models, multi-modal learning, computational biology, and precision medicine.
My path here wasn’t exactly linear: from circuit boards (undergrad in electronic engineering), to fashion (ran my own clothing store for 3 years), to data science (MS), and now to machine learning for biology. I never stop challenging myself—and that’s the fun part.
In summer 2023, I spent 6 months at Genentech, where I worked across Clinical Pharmacology and Research Biology to develop an AI model for survival prediction—integrating genomics and clinical data to guide personalized therapy.
I’m excited by big questions like:
• Can we simulate an individual’s biology before testing a drug?
• What if we could transfer knowledge from one disease to another?
• How close are we to a universal foundation model of human health?
Let’s find out 🤗.
📝 Publications
Journal Articles
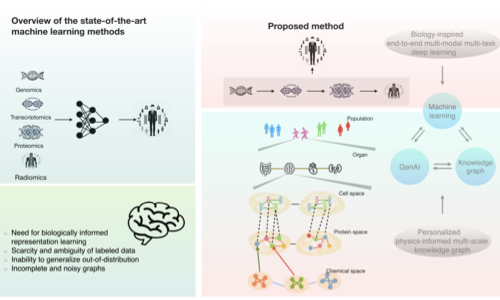
Wu, Y, Xie, L.
Computational and Structural Biotechnology Journal, 27:265-277
- Developed frameworks for integrating multi-omics data to predict complex biological phenotypes
- Applied advanced AI techniques to model genotype-environment-phenotype relationships
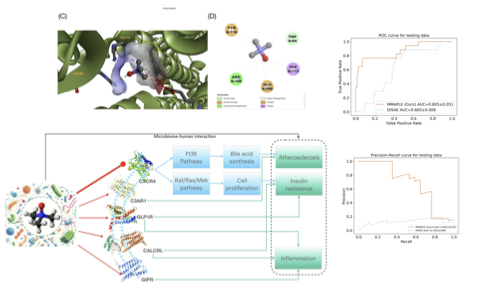
Semi-supervised meta-learning elucidates understudied molecular interactions
Wu, Y, Xie, L., Liu, Y., Xie, L.
Communications Biology, 7(1): 1104
- Developed semi-supervised meta-learning approaches to address label insufficiency in biomedical data
- Discovered novel interspecies metabolite-protein interactions
- Advanced understanding of microbiome-host interaction mechanisms
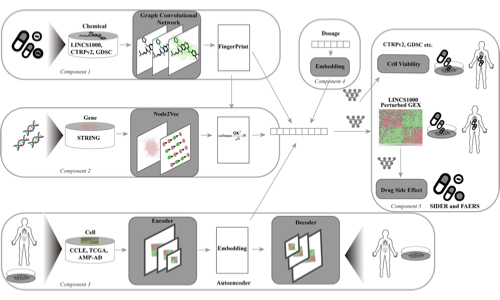
Wu, Y, Liu, Q., Xie, L.
Cell Reports Methods, 3(4)
- Integrated hierarchical multi-omics data to reconstruct proteome-scale drug-target interactions
- Built models to predict drug phenotypic responses for drug repurposing and discovery
- Focused application on incurable diseases through systems-level modeling
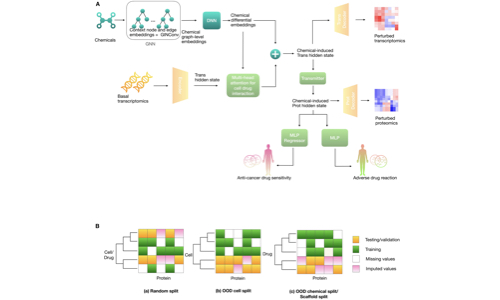
Wu, Y, Liu, Q., Qiu, Y., Xie, L.
PLoS Computational Biology, 18(8): e1010367
- Developed deep learning models for predicting complex drug responses
- Applied models to personalized Alzheimer’s disease drug repurposing
- Accounted for dose-dependent and context-specific multiplex phenotypes
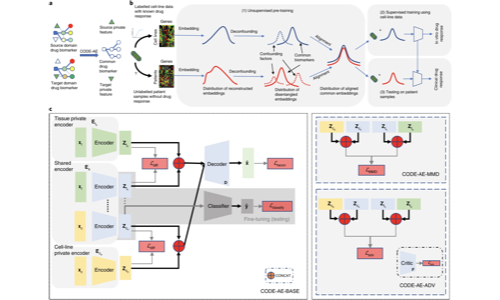
He, D., Liu, Q., Wu, Y, Xie, L.
Nature Machine Intelligence, 4(10): 879-892
- Developed a novel autoencoder architecture to handle confounding factors in drug response prediction
- Created a context-aware approach to improve personalized medicine applications
- Enhanced prediction robustness when transferring from cell-line to clinical data
COVID-19 multi-targeted drug repurposing using few-shot learning
Liu, Y., Wu, Y, Shen, X., Xie, L.
Frontiers in Bioinformatics, 1: 693177
- Applied few-shot learning techniques to the challenge of COVID-19 drug repurposing
- Developed a multi-targeted approach to identify potential therapeutic compounds
- Leveraged limited data to make predictions about drug efficacy against SARS-CoV-2
Conference Papers & Workshop Papers
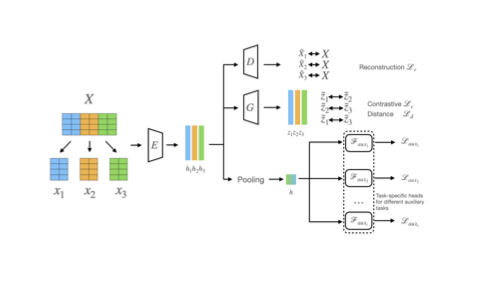
Multitask-Guided Self-Supervised Tabular Learning for Patient-Specific Survival Prediction
Wu, Y, Bazgir, O., Lee, Y., Biancalani, T., Lu, J., Hajiramezanali, E.
Neural Information Processing Systems (NeurIPS 2023): New Frontiers of AI for Drug Discovery and Development (AI4D), Table Representation Learning (TRL). Proceedings of the 18th Machine Learning in Computational Biology meeting, PMLR 240:10-22
- Developed Guided-STab, a framework using RNA-seq data pretraining across various cancer types
- Introduced novel tabular data augmentation techniques for improved representation learning
- Leveraged sparse clinical features as auxiliary multitask objectives to enhance model performance
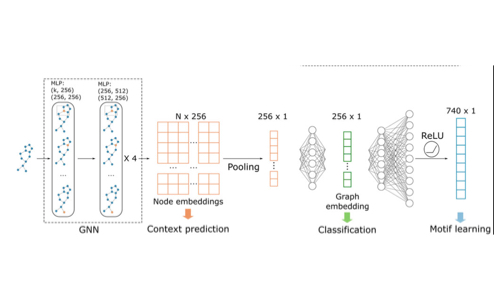
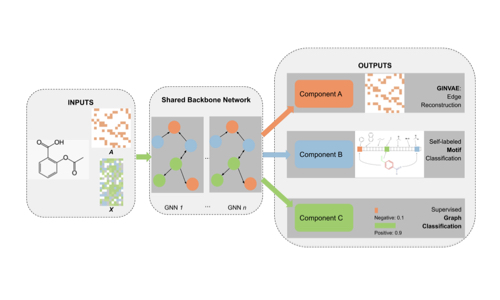
MoLGNN: Self-supervised motif learning graph neural network for drug discovery
Shen, X., Liu, Y., Wu, Y, Xie, L.
Neural Information Processing Systems (NeurIPS 2020): Machine Learning for Molecules (ml4molecules)
- Created a novel graph neural network architecture that incorporates molecular motif information
- Developed self-supervised learning strategies for molecular representation
- Applied the approach to drug discovery problems with limited labeled data
📖 Educations
- 2020 - 2025 (Expected), Ph.D. in Computer Science, City University of New York, The Graduate Center, New York, NY, USA
- Advisor: Dr. Lei Xie (City University of New York & Weill Cornell Medicine)
- Research area: Self-supervised learning (e.g., large language model), transfer learning, meta-learning, multi-modal Learning, computational biology, multi-omics data integration, drug discovery, and precision medicine
-
2020, Master of Science in Computer Information Systems (Data Science track), City University of New York, Baruch College, New York, NY, USA
- 2014, Bachelor of Engineering in Communication Engineering, University of Electronic Science and Technology of China, Chengdu, China
💬 Presentations
Invited Talks
- Fall 2024, AI-driven multi-omics integration for multi-scale predictive modeling of causal genotype-environment-phenotype relationships. Vanderbilt University Medical Center, Virtual
- Spring 2024, Harnessing AI for Omics-Driven Drug Discovery. Appel Forum WIPS, Weill Cornell Medicine, NYC
- Summer 2023, MultiDCP: A chemical perturbation deep learning modeling for dose-dependent and context-specific multiplex phenotype responses. Genentech, South San Francisco, CA
Conference & Symposium Oral Presentations
- Spring 2024, Harnessing AI for Systems Medicine of Incurable Diseases. Keystone: AI in Biomedicine, Virtual
- Summer 2022, A Context-aware De-confounding Autoencoder for Robust Prediction of Personalized Clinical Drug Response From Cell Line Compound Screening. Intelligent Systems for Molecular Biology (ISMB), Madison, WI
- Summer 2022, Deep learning prediction of chemical-induced dose-dependent and context-specific multiplex phenotype responses and its application to personalized Alzheimer’s disease drug repurposing. International Conference on Intelligent Biology and Medicine (ICIBM), Philadelphia, Penn
Conference & Symposium Poster Presentations
- Fall 2024, MMAPLE: Meta Model Agnostic Pseudo Label Learning for Understudied Out-of-distribution Molecular Interactions. International Conference on Intelligent Biology and Medicine (ICIBM), BRC Rice University in Houston, TX
- Spring 2024, AI-powered Multi-omics data integration for Systems Medicine of Incurable Diseases. DahShu Data Science Symposium, Michigan State University (MSU), MI
- Spring 2024, Spatial and single-cell multi-omics data integration and predictive modeling. Appel Poster Symposium, Weill Cornell Medicine, NYC
- Fall 2023, Multitask-Guided Self-Supervised Tabular Learning for Patient-Specific Survival Prediction. Neural Information Processing Systems (NeurIPS): New Frontiers of AI for Drug Discovery and Development (AI4D), Table Representation Learning (TRL), New Orleans, LA
🏆 Honors and Awards
- 2023, DESRES Doctoral & Postdoctoral Fellowship, D.E. Shaw Research
- 2023, 2022, Winner, Computer Science Department Poster Competition, Graduate Center, CUNY
- 2014, Excellent Student Award, University of Electronic Science and Technology of China
📋 Outreach and Professional Development
Peer Review: Nucleic Acids Research, Scientific Reports, Nature Communications, PLoS Computational Biology, Drug Discovery Today, BMC Bioinformatics, Frontiers in Immunology, ICLR-MLGenX, NeurIPS-AI4D
Professional Memberships: International Society for Computational Biology (ISCB)
🎓 Teaching Experience
- Fall 2024, Big Data Technologies, Hunter College, Teaching Assistant
- Fall 2023, Big Data Technologies, Hunter College, Instructor
👨🏫 Mentoring
- Summer 2024, Computer Science Undergraduate, Lehigh University, NSF Research Experiences for Undergraduates (REU)
- Summer 2022, Computer Science Undergraduate, Cornell University, NSF Research Experiences for Undergraduates (REU)
